API docs¶
The list below summarizes classes and functions exposed to the user, ie. imported by:
from ffnet import *
ffnet(conec[, lazy_derivative]) |
Feed-forward neural network main class. |
mlgraph(arch[, biases]) |
Creates standard multilayer network architecture. |
tmlgraph(arch[, biases]) |
Creates multilayer network full connectivity list. |
imlgraph(arch[, biases]) |
Creates multilayer architecture with independent outputs. |
savenet(net, filename) |
Dumps network to a file using cPickle. |
loadnet(filename) |
Loads network pickled previously with savenet. |
exportnet(net, filename[, name, lang, ...]) |
Exports network to a compiled language source code. |
readdata(filename, **kwargs) |
Reads arrays from ASCII files. |
pikaia(ff, n[, ff_extra_args, individuals, ...]) |
Pikaia version 1.2 - genetic algorithm based optimizer. |
Architecture generators¶
-
ffnet.mlgraph(arch, biases=True)[source]¶ Creates standard multilayer network architecture.
Parameters: - arch : tuple
Tuple of integers - numbers of nodes in subsequent layers.
- biases : bool, optional
Indicates if bias (node numbered 0) should be added to hidden and output neurons. Default is True.
Returns: - conec : list
List of tuples – network connections.
Examples: Basic calls:
>>> from ffnet import mlgraph >>> mlgraph((2,2,1)) [(1, 3), (2, 3), (0, 3), (1, 4), (2, 4), (0, 4), (3, 5), (4, 5), (0, 5)] >>> mlgraph((2,2,1), biases = False) [(1, 3), (2, 3), (1, 4), (2, 4), (3, 5), (4, 5)]
Exemplary plot:
from ffnet import mlgraph, ffnet import networkx as NX import pylab conec = mlgraph((3,6,3,2), biases=False) net = ffnet(conec) NX.draw_graphviz(net.graph, prog='dot') pylab.show()
(Source code, png, hires.png, pdf)

-
ffnet.tmlgraph(arch, biases=True)[source]¶ Creates multilayer network full connectivity list.
Similar to
mlgraph, but now layers are fully connected with all preceding layers.Parameters: - arch : tuple
Tuple of integers - numbers of nodes in subsequent layers.
- biases : bool, optional
Indicates if bias (node numbered 0) should be added to hidden and output neurons. Default is True.
Returns: - conec : list
List of tuples – network connections.
Examples: Basic calls:
>>> from ffnet import tmlgraph >>> tmlgraph((2,2,1)) [(0, 3), (1, 3), (2, 3), (0, 4), (1, 4), (2, 4), (0, 5), (1, 5), (2, 5), (3, 5), (4, 5)] >>> tmlgraph((2,2,1), biases = False) [(1, 3), (2, 3), (1, 4), (2, 4), (1, 5), (2, 5), (3, 5), (4, 5)]
Exemplary plot:
from ffnet import tmlgraph, ffnet import networkx as NX import pylab conec = tmlgraph((3, 8, 3), biases=False) net = ffnet(conec) NX.draw_graphviz(net.graph, prog='dot') pylab.show()
(Source code, png, hires.png, pdf)

-
ffnet.imlgraph(arch, biases=True)[source]¶ Creates multilayer architecture with independent outputs.
This function uses
mlgraphto build independent multilayer architectures for each output neuron. Then it merges them into one graph with common input nodes.Parameters: - arch : tuple
Tuple of length 3. The first element is number of network inputs, last one is number of outputs and the middle one is interpreted as the hidden layers definition (it can be an integer or a list – see examples)
- biases : bool, optional
Indicates if bias (node numbered 0) should be added to hidden and output neurons. Default is True.
Returns: - conec : list
List of tuples – network connections.
Raises: - TypeError
If arch cannot be properly interpreted.
Examples: The following arch definitions are possible:
>>> from ffnet import imlgraph >>> arch = (2, 2, 2) >>> imlgraph(arch, biases=False) [(1, 3), (2, 3), (1, 4), (2, 4), (3, 5), (4, 5), (1, 6), (2, 6), (1, 7), (2, 7), (6, 8), (7, 8)]
In this case two multilayer networks (for two outputs) of the architectures (2,2,1), (2,2,1) are merged into one graph.
>>> arch = (2, [(2,), (2,2)], 2) >>> imlgraph(arch, biases=False) [(1, 3), (2, 3), (1, 4), (2, 4), (3, 5), (4, 5), (1, 6), (2, 6), (1, 7), (2, 7), (6, 8), (7, 8), (6, 9), (7, 9), (8, 10), (9, 10)]
I this case networks of the architectures (2,2,1) and (2,2,2,1) are merged.
Exemplary plot:
from ffnet import imlgraph, ffnet import networkx as NX import pylab conec = imlgraph((3, [(3,), (6, 3), (3,)], 3), biases=False) net = ffnet(conec) NX.draw_graphviz(net.graph, prog='dot') pylab.show()
(Source code, png, hires.png, pdf)
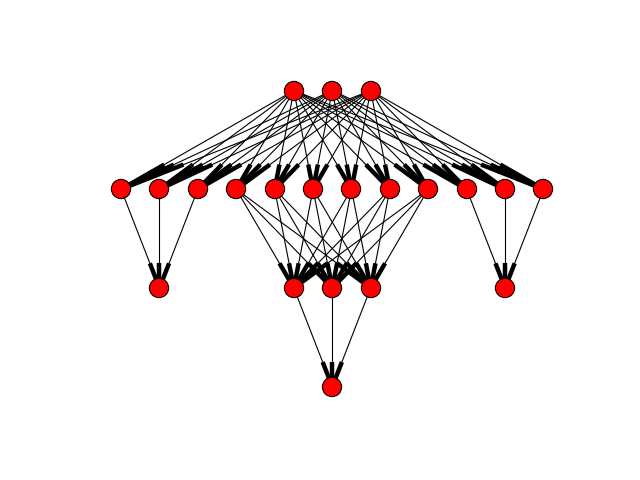
Main ffnet class¶
-
class
ffnet.ffnet(conec, lazy_derivative=True)[source]¶ Feed-forward neural network main class.
Parameters: - conec : list of tuples
List of network connections
- lazy_derivative : bool
If True all data necessary for derivatives calculation (see
ffnet.derivativemethod) are generated only on demand.
Returns: - net
Feed forward network object
Raises: - TypeError
If conec is not directed acyclic graph
Instance attributes: - conec : array
Topologically sorted network connections
- weights : array
Weights in order of topologically sorted connections
- renormalize : bool
If True normalization ranges will be recreated from training data at next training call.
Default is True
Examples: >>> from ffnet import mlgraph, ffnet >>> conec = mlgraph((2,2,1)) >>> net = ffnet(conec)
See also: -
call(inp)[source]¶ Calculates network answer to given input.
Parameters: - inp : array
2D array of input patterns (or 1D for single pattern)
Returns: - ans : array
1D or 2D array of calculated network outputs
Raises: - TypeError
If inp is invalid
-
derivative(inp)[source]¶ Returns partial derivatives of the network’s output vs its input.
For each input pattern an array of the form:
| o1/i1, o1/i2, ..., o1/in | | o2/i1, o2/i2, ..., o2/in | | ... | | om/i1, om/i2, ..., om/in |
is returned.
Parameters: - inp : array
2D array of input patterns (or 1D for single pattern)
Returns: - ans : array
1D or 2D array of calculated network outputs
Examples: >>> from ffnet import mlgraph, ffnet >>> conec = mlgraph((3,3,2)) >>> net = ffnet(conec); net.weights[:] = 1. >>> net.derivative([0., 0., 0.]) array([[ 0.02233658, 0.02233658, 0.02233658], [ 0.02233658, 0.02233658, 0.02233658]])
-
randomweights()[source]¶ Randomize network weights due to Bottou proposition.
If n is a number of incoming connections to the node, weights of these connections are chosen randomly from range (-2.38/sqrt(n), 2.38/sqrt(n))
-
sqerror(input, target)[source]¶ Calculates sum of squared errors at network output.
Error is calculated for normalized input and target arrays.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
Returns: - err : float
0.5*(sum of squared errors at network outputs)
Note
This function might be slow in frequent use, because data normalization is performed at each call. Usually there’s no need to use this function, unless you need to adopt your own training strategy.
-
sqgrad(input, target)[source]¶ Returns gradient of network error vs. network weights.
Error is calculated for normalized input and target arrays.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
Returns: - grad : 1-D array
Array of the same length as net.weights containing gradient values.
Note
This function might be slow in frequent use, because data normalization is performed at each call. Usually there’s no need to use this function, unless you need to adopt your own training strategy.
-
test(input, target, iprint=1, filename=None)[source]¶ Calculates output and parameters of regression.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
- iprint : {0, 1, 2}, optional
Verbosity level: 0 – print nothing, 1 – print regression parameters for each output node (default), 2 – print additionaly general network info and all targets vs. outputs
- filename : str
Path to the file where printed messages are redirected Default is None
Returns: - out : tuple
(output, regress) tuple where: output is an array of network answers on input patterns and regress contains regression parameters for each output node. These parameters are: slope, intercept, r-value, p-value, stderr-of-slope, stderr-of-estimate.
Examples: >>> from ffnet import mlgraph, ffnet >>> from numpy.random import rand >>> conec = mlgraph((3,3,2)) >>> net = ffnet(conec) >>> input = rand(50,3); target = rand(50,2) >>> output, regress = net.test(input, target) Testing results for 50 testing cases: OUTPUT 1 (node nr 8): Regression line parameters: slope = -0.000649 intercept = 0.741282 r-value = -0.021853 p-value = 0.880267 slope stderr = 0.004287 estim. stderr = 0.009146 . OUTPUT 2 (node nr 7): Regression line parameters: slope = 0.005536 intercept = 0.198818 r-value = 0.285037 p-value = 0.044816 slope stderr = 0.002687 estim. stderr = 0.005853
Exemplary plot:
from ffnet import mlgraph, ffnet from numpy.random import rand from numpy import linspace import pylab # Create and train net on random data conec = mlgraph((3,10,2)) net = ffnet(conec) input = rand(50,3); target = rand(50,2) net.train_tnc(input, target, maxfun = 400) output, regress = net.test(input, target, iprint = 0) # Plot results for first output pylab.plot(target.T[0], output.T[0], 'o', label='targets vs. outputs') slope = regress[0][0]; intercept = regress[0][1] x = linspace(0,1) y = slope * x + intercept pylab.plot(x, y, linewidth = 2, label = 'regression line') pylab.legend() pylab.show()
(Source code, png, hires.png, pdf)

-
train_bfgs(input, target, **kwargs)[source]¶ Train network with constrained version of BFGS algorithm.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
- maxfun : int
Maximum number of function evaluations (default is 15000)
- bounds : list, optional
(min, max) pairs for each connection weight, defining the bounds on that weight. Use None for one of min or max when there is no bound in that direction. By default all bounds ar set to (-100, 100)
- disp : int, optional
If 0, then no output (default). If positive number then convergence messages are dispalyed.
See also
scipy.optimize.fmin_l_bfgs_boptimizer is used in this method. Look at its documentation for possible other useful parameters.
-
train_cg(input, target, **kwargs)[source]¶ Train network with conjugate gradient algorithm.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
- maxiter : integer, optional
Maximum number of iterations (default is 10000)
- disp : bool
If True convergence method is displayed (default)
See also
scipy.optimize.fmin_cgoptimizer is used in this method. Look at its documentation for possible other useful parameters.
-
train_genetic(input, target, **kwargs)[source]¶ Global weights optimization with genetic algorithm.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
- lower : float, optional
Lower bound of weights values (default is -25.)
- upper : float, optional
Upper bound of weights values (default is 25.)
- individuals : integer, optional
Number of individuals in a population (default is 20)
- generations : integer, optional
Number of generations over which solution is to evolve (default is 500)
- verbosity : {0, 1, 2}, optional
Printed output 0/1/2=None/Minimal/Verbose (default is 0)
See also
See description of
pikaia.pikaiaoptimization function for other parameters.
-
train_momentum(input, target, eta=0.2, momentum=0.8, maxiter=10000, disp=0)[source]¶ Simple backpropagation training with momentum.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
- eta : float, optional
Learning rate
- momentum : float, optional
Momentum coefficient
- maxiter : integer, optional
Maximum number of iterations
- disp : bool
If True convergence method is displayed
-
train_rprop(input, target, a=1.2, b=0.5, mimin=1e-06, mimax=50.0, xmi=0.1, maxiter=10000, disp=0)[source]¶ Rprop training algorithm.
Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
- a : float, optional
Training step increasing parameter
- b : float, optional
Training step decreasing parameter
- mimin : float, optional
Minimum training step
- mimax : float, optional
Maximum training step
- xmi : array (or float), optional
Array containing initial training steps for weights. If xmi is a scalar then its value is set for all weights
- maxiter : integer, optional
Maximum number of iterations
- disp : bool
If True convergence method is displayed. Default is False
Returns: - xmi : array
Computed array of training steps to be used in eventual further training calls.
-
train_tnc(input, target, nproc=1, **kwargs)[source]¶ Parameters: - input : 2-D array
Array of input patterns
- target : 2-D array
Array of network targets
- nproc : int or ‘ncpu’, optional
Number of processes spawned for training. If nproc=’ncpu’ nproc will be set to number of avilable processors
- maxfun : int
Maximum number of function evaluation. If None, maxfun is set to max(100, 10*len(weights)). Defaults to None.
- bounds : list, optional
(min, max) pairs for each connection weight, defining the bounds on that weight. Use None for one of min or max when there is no bound in that direction. By default all bounds ar set to (-100, 100)
- messages : int, optional
If 0, then no output (default). If positive number then convergence messages are dispalyed.
Note
On Windows using ncpu > 1 might be memory hungry, because each process have to load its own instance of network and training data. This is not the case on Linux platforms.
See also
scipy.optimize.fmin_tncoptimizer is used in this method. Look at its documentation for possible other useful parameters.
Utility functions¶
-
ffnet.savenet(net, filename)[source]¶ Dumps network to a file using cPickle.
Parameters: - net : ffnet
Intance of the network
- filename : str
Path to the file where network is dumped
-
ffnet.loadnet(filename)[source]¶ Loads network pickled previously with
savenet.Parameters: - filename : str
Path to the file with saved network
-
ffnet.exportnet(net, filename, name='ffnet', lang=None, derivative=True)[source]¶ Exports network to a compiled language source code.
Parameters: - filename : str
Path to the file where network is exported
- name : str
Name of the exported function
- lang : str
Language to which network is to be exported. Currently only Fortran is supported
- derivative : bool
If True a function for derivative calculation is also exported. It is named as name with prefix ‘d’
Note
You need ‘ffnet.f’ file distributed with ffnet sources to get the exported Fortran routines to work.
Pikaia optimizer¶
-
pikaia.pikaia(ff, n, ff_extra_args=(), individuals=100, generations=500, digits=6, crossover=0.85, mutation=2, initrate=0.005, minrate=0.0005, maxrate=0.25, fitnessdiff=1.0, reproduction=3, elitism=0, verbosity=0)[source]¶ Pikaia version 1.2 - genetic algorithm based optimizer.
Simplest usage:
from pikaia import pikaia x = pikaia(ff, n)
Parameters: - ff : callable
Scalar function of the signature ff(x, [n, args]), where x is a real array of length n and args are extra parameters. Pikaia optimizer assumes x elements are bounded to the interval (0, 1), thus ff have to aware of this, ie. probably you need some internal scaling inside ff.
By convention, ff should return higher values for more optimal parameter values (i.e., individuals which are more “fit”). For example, in fitting a function through data points, ff could return the inverse of chi**2.
- n : int
Length of x. Note that you do not need starting point.
- ff_extra_args: tuple
Extra arguments passed to ff.
- individuals : int
Number of individuals in a population (default is 100)
- generations : int
Number of generations over which solution is to evolve (default is 500)
- digits : int
Number of significant digits (i.e., number of genes) retained in chromosomal encoding (default is 6). (Note: This number is limited by the machine floating point precision. Most 32-bit floating point representations have only 6 full digits of precision. To achieve greater precision this routine could be converted to double precision, but note that this would also require a double precision random number generator, which likely would not have more than 9 digits of precision if it used 4-byte integers internally.)
- crossover : float
Crossover probability; must be <= 1.0 (default is 0.85). If crossover takes place, either one or two splicing points are used, with equal probabilities.
- mutation : {1, 2, 3, 4, 5, 6}
digit description 1 one-point mutation, fixed rate 2 one-point, adjustable rate based on fitness (default) 3 one-point, adjustable rate based on distance 4 one-point+creep, fixed rate 5 one-point+creep, adjustable rate based on fitness 6 one-point+creep, adjustable rate based on distance - initrate : float
Initial mutation rate. Should be small (default is 0.005) (Note: the mutation rate is the probability that any one gene locus will mutate in any one generation.)
- minrate : float
Minimum mutation rate. Must be >= 0.0 (default is 0.0005)
- maxrate : float
Maximum mutation rate. Must be <= 1.0 (default is 0.25)
- fitnessdiff : float
Relative fitness differential. Range from 0 (none) to 1 (maximum) (default is 1).
- reproduction : {1, 2, 3}
Reproduction plan; 1/2/3 = Full generationalreplacement/Steady- state-replace-random/Steady-state-replace-worst (default is 3)
- elitism : {0, 1}
Elitism flag; 0/1=off/on (default is 0) (Applies only to reproduction plans 1 and 2)
- verbosity : {0, 1, 2}
Printed output 0/1/2=None/Minimal/Verbose (default is 0)
Returns: - x : array (float32)
The ‘fittest’ (optimal) solution found, i.e., the solution which maximizes fitness function ff.
Examples: >>> from pikaia import pikaia >>> def ff(x): return -sum(x**2) >>> pikaia(ff, 4, individuals=50, generations=200) array([ 1.23000005e-04, 7.69999970e-05, 2.99999992e-05, 2.80000004e-05], dtype=float32)
Note
Original fortran code of pikaia is written by: Paul Charbonneau & Barry Knapp (paulchar@hao.ucar.edu, knapp@hao.ucar.edu)
Wrapped with f2py by Marek Wojciechowski (mwojc@p.lodz.pl)
